Fragment-based hit discovery for the epigenetic target, BRD3
Fragment-based drug discovery allows a wide section of chemical space to be interrogated with a relatively small but diverse library of small molecules.
Fragment-based approaches have been applied successfully to the discovery and optimization of ligands for a wide range of molecular targets, including epigenetic targets. Sygnature Discovery has designed and synthesised a proprietary fragment library with a high degree of novelty that covers a wide range of chemical space. Comprised of over 1,000 compounds, most of which are not commercially available, the library was designed using the ‘rule of 3’ as a guideline, with a strong emphasis on diversity. It is therefore readily amenable to medicinal chemistry optimisation.
Implementation
Figure 1: PDB rendering of 2XNB
To demonstrate the synergy between Sygnature Discovery and Peak Proteins in the arena of fragment-based drug discovery, Sygnature’s proprietary fragment library was screened against an epigenetic target with little previous literature on drug discovery, bromodomain-containing protein 3 (BRD3) (Figure 1). Bromodomain-containing proteins function can affect the transcription of DNA. BRD3 functions by binding acetylated lysine residues on chromatin and transcriptional regulators.
Due to this involvement in regulation, Bromodomain and Extra-terminal motif (BET) members often play a role in several types of cancer and in some metabolic disorders. BRD3 in particular is associated with a number of disease phenotypes, for example, depletion of BRD3 slows growth in cancer models including prostate cancer and medulloblastoma. BRD3 has also been implicated in NUT midline carcinoma (NMT).
While there are a number of available pan-BET inhibitors for cancer treatment, the design of selective inhibitors may lead to a more beneficial clinical outcome due to reduced off-target effects. BRD3 therefore represents an intriguing and challenging drug discovery target.
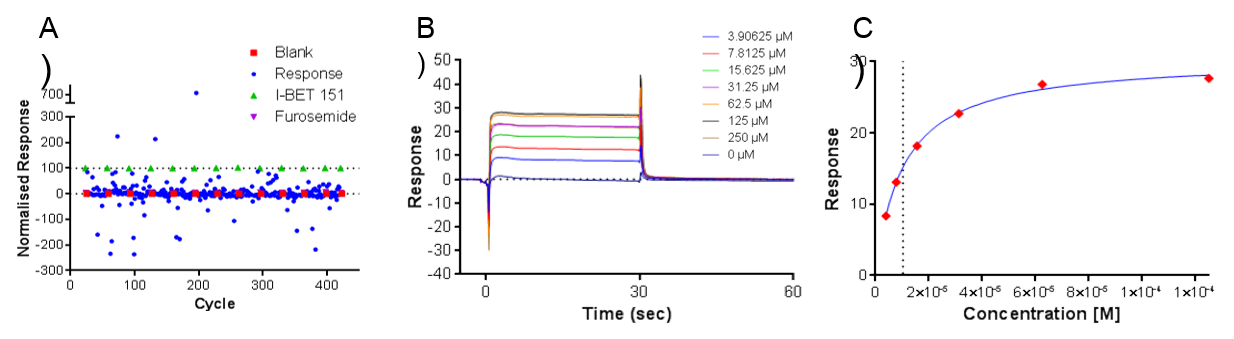
Figure 2. A) Example SPR data showing relative responses across 320 fragments, B) Response to concentration curve of fragment and C) Fit of steady state data
Surface Plasmon Resonance (SPR)
Surface plasmon resonance (SPR) is a biophysical technique which allows an assessment of both the kinetics and affinity of a compound binding to an immobilised protein surface. Using SPR, Sygnature’s proprietary fragment library and BRD3 protein expressed in E. coli and purified by Peak Proteins.
In total, 908 fragments were screened for binding to the first domain of BRD3 (BRD3D1) as well as for selectivity against other bromodomain and unrelated protein target surfaces. Of the fragments screened, nine were found to bind BRD3 with affinities ranging from <10 µM to 250 µM (Figure 2). Peak Proteins then confirmed the binding of the fragments by X-ray crystallography at the Diamond Light Source (Oxford, UK).
Of the nine fragments analysed, six were found to contain bound fragments, with X-ray crystallographic structures resolved to 1.4 to 1.7 Å. All six were confirmed from the structures to bind in the expected Ac-Lys, as exemplified in Figure 3. By using these fragments as starting points to develop tool ligands, we could further interrogate BRD3 involvement in disease phenotypes, as well as using these as potential starting points for a new drug discovery programme.
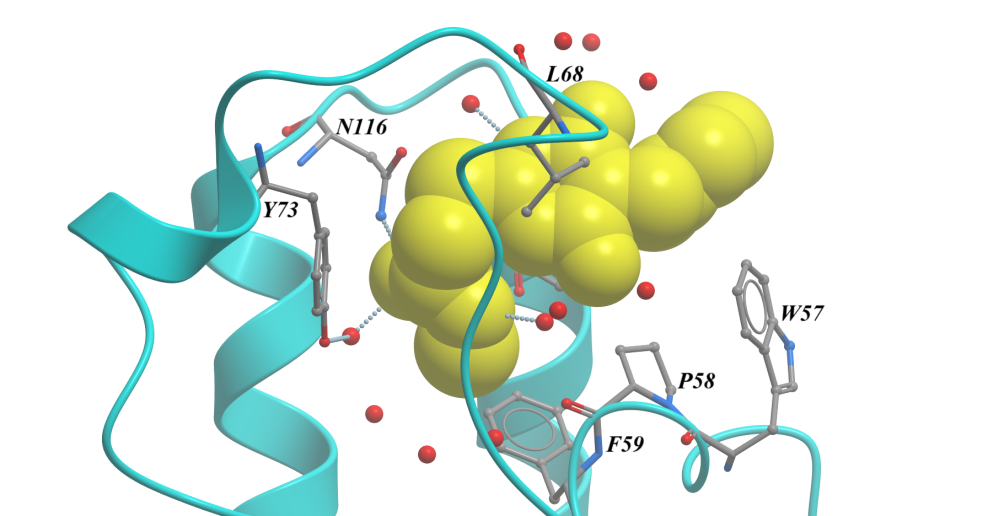
Figure 3. BRD3D1 structure bound to SYG-00000127, 1.69 Å. <10 µM KD by SPR.
