Enhancing Membrane Protein Characterisation: Sygnature, Peak Proteins & Cytiva Collaboration
Advancing the biophysical characterisation of membrane protein targets in drug discovery: A collaboration amongst Sygnature Discovery, Peak Proteins and Cytiva
We’re pleased to announce a new collaboration between protein scientists at Peak Proteins (now a Sygnature Company), members of the biophysics team at Sygnature Discovery, and surface plasmon resonance (SPR) experts at Cytiva to develop methods for membrane protein analysis. The project aims to assess and develop techniques using polymer encapsulated G-protein coupled receptors (GPCRs) and other membrane proteins for SPR studies of small-molecule binding.
Cookie-cutters – the secret to alleviating the bottlenecks in working with membrane protein targets?
30% of all pharmaceuticals target GPCRs. With the challenges associated with the production of soluble and stable GPCR protein as a reagent for drug discovery, there is a need for technological advances to help solve this bottleneck.
Traditionally, the study of isolated membrane proteins has relied on solubilisation from the membrane in detergent micelles. However, detergent solubilisation has a number of drawbacks, namely that the majority of native lipids are stripped from the protein, often resulting in an unstable or inactive protein product. Sounds challenging, right?
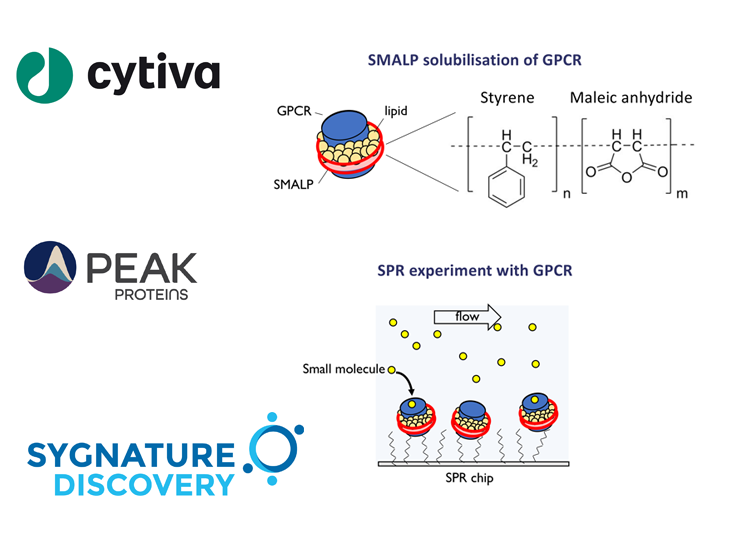
Alternatively, there are a growing number of detergent-free technologies that allow the extraction of membrane proteins without losing native lipids. One such technology is the use of styrene maleic acid (SMA) co-polymers that can self-assemble in membrane bi-layers and act as ‘cookie-cutters’ to excise protein/lipid discs known as SMA lipid particles (SMALPs) from the membrane.
Membrane proteins extracted in SMALP or other co-polymers could hold a number of key advantages for biophysical characterisation by SPR analysis. For example, by using SMALP encapsulated protein, a prolonged stability of GPCR on sensor chip surface, the ability to incubate at higher temperatures during the assay, and a more native-like environment with lipids rather than in a detergent micelle would be expected. This all translates to facilitating the ability to screen and characterise modulators of membrane protein targets in a drug discovery program.
Building on established methodology
Characterisation of protein:protein interaction between SMALP encapsulated membrane protein and antibody binding partner has already been accomplished. Using SMALP-encapsulated membrane protein for protein:small-molecule interaction analysis is a more challenging prospect. The protein in SMALP is predicted to be a relatively large complex, therefore high sensitivity instrumentation and high quality protein are likely requirements of a successful assay. Another technical challenge is the choice of immobilisation method; the pH sensitive nature of these polymers combined with their negative charge makes a classical covalent coupling method challenging. Capture methods are likely to be more successful but come with their own issues, including the occlusion of tags and other charge-based interference.
Using a number of tricks and careful assay development, we hope to overcome these challenges in order to optimise a method for using SMALP encapsulated GPCRs in SPR for protein:small-molecule interaction analysis.
The recipe for success
Steve Harborne is leading activities at Peak Proteins, producing purified GPCRs in SMALP and other detergent-free co-polymers for analysis. Ella Halcrow from Sygnature Discovery has been developing Biacore SPR and other biophysical assays using the isolated GPCRs supported by Philip Addis and Scott Pollack. Cytiva is providing SPR consumable support as well as technical expertise from Anna Moberg, Gillian Goodwin and Claire Shepherd.
About the author
Steve Harborne is a membrane protein specialist at Peak Proteins (a Sygnature company) with expertise in a range of protein expression and purification techniques in particular applied to challenging membrane protein targets.
If you would like to know more about biophysics and its applications for tackling membrane protein targets, contact our team by using the form below.
