Executing hit-finding against a novel nucleic acid binding complex – a structurally enabled approach identifying hits against different complex states
Well-characterized hits provide a strong foundation for experienced drug discovery teams to rapidly advance a compound to the candidate stage. In our #StrongFoundations series, the Sygnature Discovery team takes you through three successful hit ID case studies which demonstrate how to get the best start points for your projects.
Challenge
Nucleic acid binding proteins are involved in a wide range of fundamental processes, making members of this class desirable for therapeutic targeting. A challenge with these complexes is their dynamic nature – often the co-ordination of the nucleic acid substrate can be accompanied by dynamic changes within the structure, meaning structural data is not always available for all forms.
For the target in question, structural data was available for a truncated form of the target domain bound to the nucleic acid substrate but not in the absence of substrate. As is common in these families, the binding of the nucleic acid substrate confers a more rigid confirmation.
Approach: Using DSF to profile hits against bound vs. unbound target
To understand how valid this structural information was to progress future hit series, a focused crystallography screen was conducted in parallel with an extensive differential scanning fluorimetry (DSF) also called TSA or FTSA (fluorescent thermal shift assay) profiling programme. The aim was to identify to which form of the target hits were found: bound vs unbound and whether there was overlap between hits against the full length target and the truncated crystallography construct.
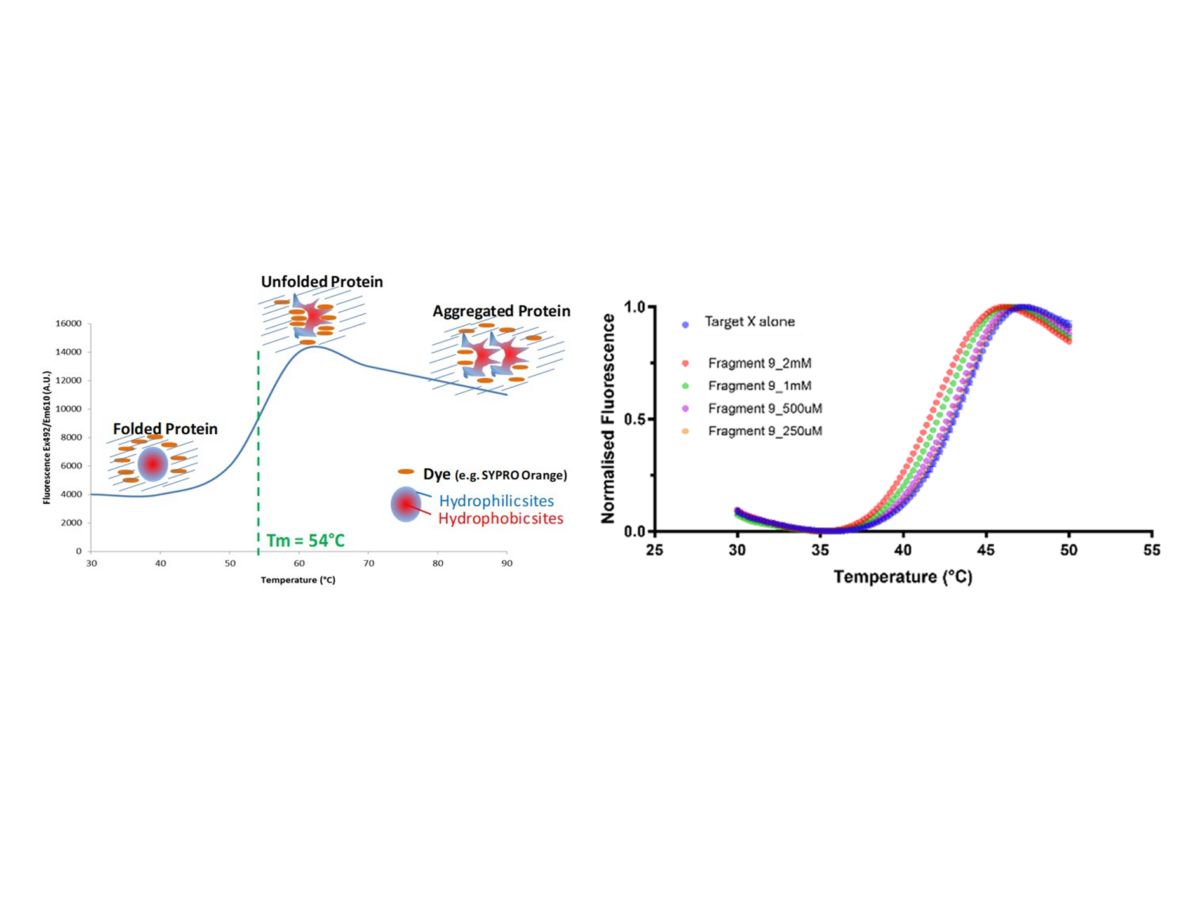
Figure 1: Illustration of the fluorescent responses observed as a protein unfolds in a differential scanning fluorimetry experiment (L), data from a DSF experiment for target alone in the presence of varying concentrations of a hit fragment, illustrating the effect on the target melting temperature, Tm (R).
Hit profiling
Using DSF a full screen of the Sygnature Fragment Library under the following conditions:
- FL target X
- FL target X + substrate
- Truncated target X
- Truncated target X + substrate
The output from these screens was extensively profiled to characterise different cohorts within the identified hits as shown in Figure 2:
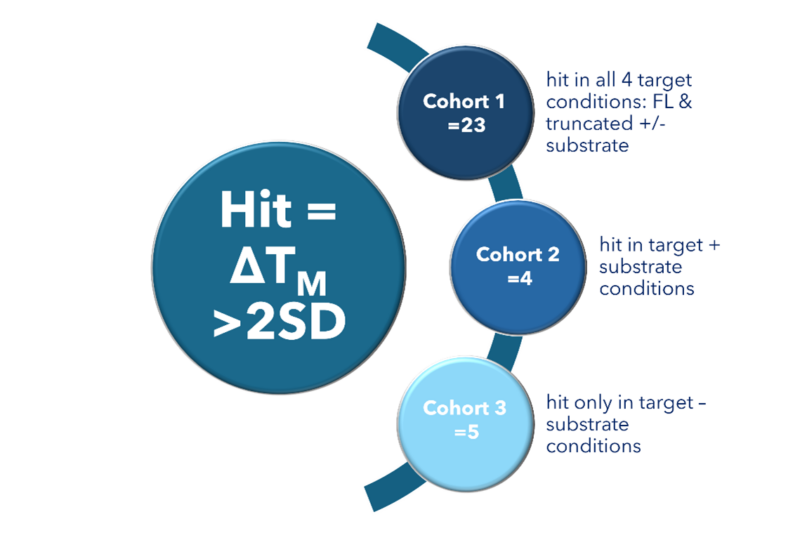
Figure 2: Analysis of the DSF screening outputs was undertaken to understand how the fragments from the Sygnature Fragment Library affected the Tm for the target protein as FL vs. truncated and +/- substrate. This analysis identified different cohorts of hits and subsequently three cohorts were categorised for follow-up based on the target complex state they bound.
In parallel, a focused sub-deck from the Sygnature Fragment Library was also screened by crystallography. This was a challenging screen, requiring crystal handling at 4oC, however, after optimization of the crystallisation process, this resulted in a number of structures that supported the DSF results.
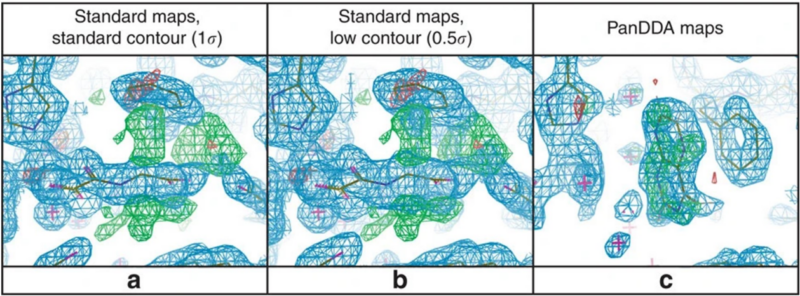
Figure 3: PanDDA maps clearly show detail obscured by conventional maps; PanDDA example from Pearce et al (2017) showing fragment screening X-ray data at 1.48 Å. (a,b) Conventional electron density maps (2mFo –DFc, contoured in blue & mFo –DFc, in green/red, ±3σ) are very difficult to interpret, dominated by a co-factor analogue bound in the majority fraction of the crystal, whereas (c) the PanDDA event map unambiguously reveals both the bound fragment and associated changes in protein conformation.
Screening outcomes and understanding the hits
There were several distinct cohorts that could be identified from the DSF library screening outputs, as shown in Figure 2, this illustrated the different characteristics of the full length and truncated target as well as +/- nucleic acid substrate binding.
From this breakdown of the screening output, three cohorts were selected as priority for follow-up. Cohort 1 designated the 23 fragments that were confirmed to have some interaction with the target in all 4 conditions, 2 of these were also identified from the crystallography focused selection, giving us early confidence in this set. This cohort was particularly interesting, since these hits bound both the substrate bound and unbound target, suggesting that some regions of the target at least must be minimally affected by substrate binding, indicating a common binding site available in both complexes. This might also imply these hits did not prevent substrate binding; however, it was not believed at this stage that this was required to inhibit function. It has since been confirmed these hits demonstrate functional inhibition. Furthermore, since these hits must have binding sites within the truncated construct, we would be able to drive future medicinal chemistry development on this hit matter using the crystallography derived structural data.
Two additional cohorts were flagged for later investigation: cohort 2 was the cohort of hits that bound both constructs only with the substrate bound and the opposing scenario was covered by cohort 3, hits that bound both constructs only in the absence of the substrate. Within these categories we had provided a diverse strategy of options for the future programme development, if the cohort 1 hits had not demonstrated functional inhibition, we had two further cohorts which would enable an alternative approach.
Conclusion
Overall, this robust and informed hit-finding strategy gave the project a strong foundation from which to initiate the hit-to-lead stage.
While sometimes dismissed as a simplistic platform with a binary data output, DSF was a driving force in this programme and allowed a tunability to the assay setup that could not achieved with other perceived “higher” value platforms. With the DSF data we were able to build confidence in the available structural data being applicable for development of a number of the hit cohorts we had identified. Furthermore, the DSF platform enabled screening of a larger number of compounds than would have been feasible using the low temperature crystallography system alone.
A multi-assay approach to fully inform hit-finding is empowering when working with challenging targets and is often necessary when the “ideal” scenario cannot be achieved in the preferred screening platform.
PanDDA reference:
Pearce, N., Krojer, T., Bradley, A. et al. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat Commun 8, 15123 (2017). https://doi.org/10.1038/ncomms15123
Find out more about Sygnature Discovery’s powerful hit ID platform, HIT SYNERGY, which integrates multiple advanced screening technologies to rapidly generate high-quality, structurally diverse hits, tailored to today’s most challenging new target classes. Well-characterized hits provide a strong foundation for our experienced drug discovery teams to advance your compound to the candidate stage.
