Biophysics: How to choose the right assay for your drug discovery project
Outside of drug discovery and some academic groups, biophysics is a relative unknown. This branch of bioscience focuses on the analysis and quantitation of biomolecules using physics, often generating data that cannot be produced by other means. It primarily focuses on assessing candidate drug-target binding strength and answering crucial questions about potency, duration, and selectivity.
Biophysical assays are also significantly less complex than cellular assays, meaning that an interaction can be assessed without interference from other factors, however, this can both be an advantage and a disadvantage. Multiple components can also be used if required, helping to determine mechanisms of action for the potential therapeutics.
We firmly believe that integrating biophysics into the discovery process enhances our comprehension of drugs and guides projects effectively when combined with other assay systems.
What Biophysical Data Does a Project Need?
 The immediate response might be “gather as much data as possible!” While partly true, the focus should be on generating the correct data for a project. If the correct data isn’t generated a project cannot progress, wasting time and resources, which may be better spent elsewhere.
The immediate response might be “gather as much data as possible!” While partly true, the focus should be on generating the correct data for a project. If the correct data isn’t generated a project cannot progress, wasting time and resources, which may be better spent elsewhere.
So, what is the correct data?
This varies on the target of the drug molecule itself, the modality of the drug (how do you expect it to work), the type of drug molecule (small molecule vs. biologic), or the disease indication to be treated. It also depends very heavily on the stage of the project. For example, it is rarely worth trying to gather detailed kinetic information during a hit-finding process, where affinities are low, and validation of target binding and behaviour is key. Similarly, yes/no target engagement data is often not required at the later stages of the project, where information on residence time, behaviour and mechanism of action are much more critical.
It is essential to continually assess the state of the project and what the key questions are to help move things forward. This is true of biophysics in the same way as for all aspects of a discovery project.
Common Biophysics Platforms and Their Advantages
How can biophysics help, and what technologies are available? This discussion primarily focuses on well-established biophysical techniques, it’s important to note that a wide array of specialised methods exists, each suited to specific applications. We are seeing a noticeable shift towards cell-based biophysical technologies as new techniques are developed, but many of these are still in their infancy and, barring CETSA® and its related techniques are yet to be widely adopted throughout the industry.
The most commonly used biophysical techniques include:
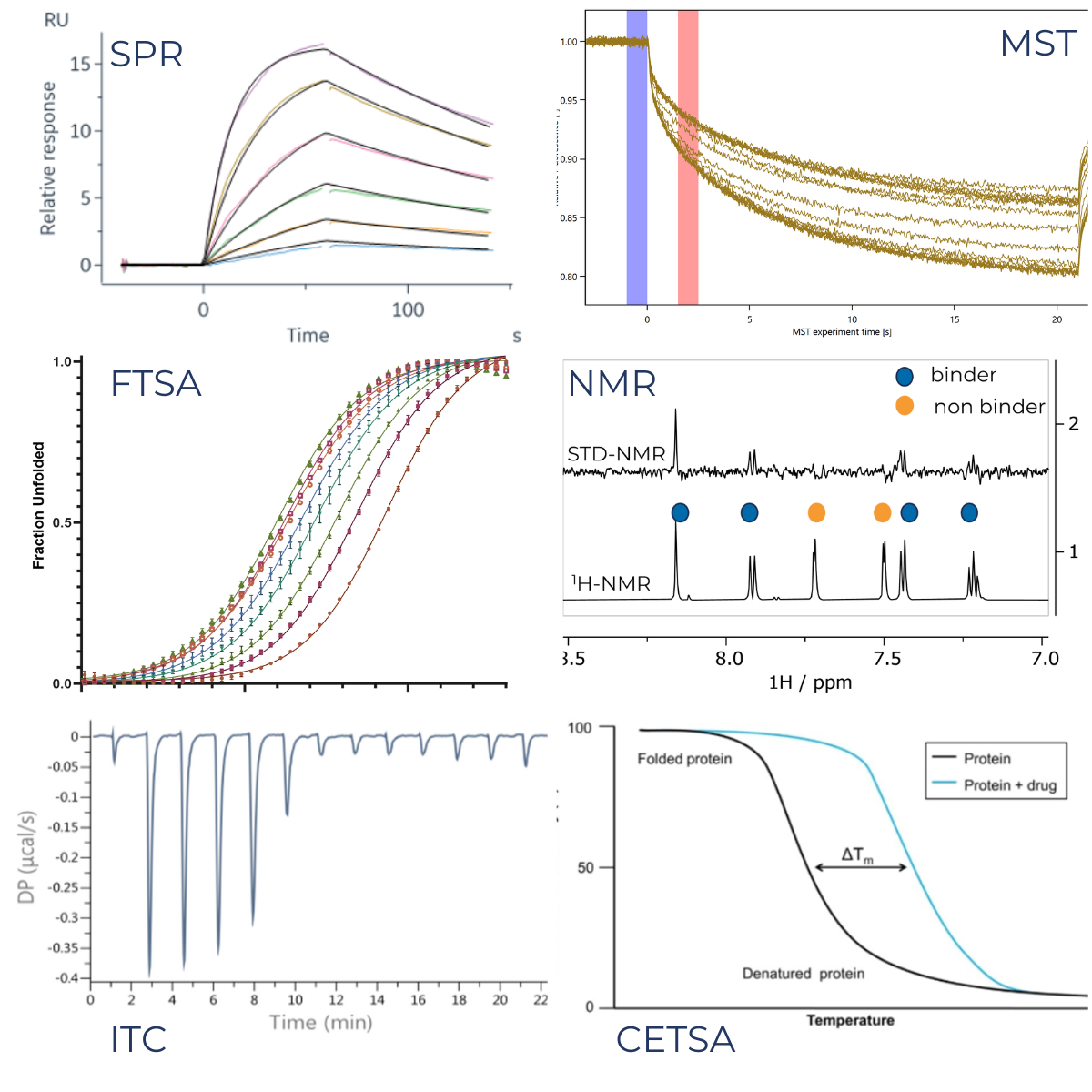
- Surface Plasmon Resonance (SPR)
- Microscale Thermophoresis (MST)
- Nuclear Magnetic Resonance (NMR)
- Isothermal Titration Calorimetry (ITC)
- Fluorescence Thermal Shift Assays (FTSA, also called Differential scanning Fluorimetry or DSF)
- Cellular thermal shift assays (CETSA®)
These technologies form the core of most biophysical offerings and have various advantages and disadvantages relative to each other. Due to this, they are suited to different applications and can answer different questions depending on the requirements of a project. Access to this toolbox progresses projects and generates the data that a project needs.
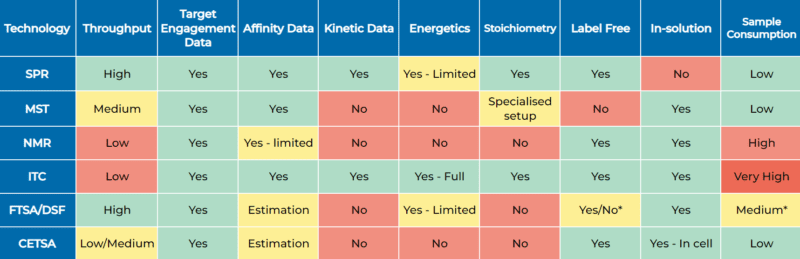
*FTSA/DSF assay sensitivity and label options depend on setup. Intrinsic tryptophan fluorescence-based DSF techniques are label-free and may have better sensitivity than dye-based assays in some situations.
Biophysical assays excel in target engagement assessment, a core strength shared by all these technologies. They also have their additional characteristics. For example, SPR is the gold standard for generating kinetic data and is highly flexible regarding assay setup. It is also the highest throughput of the techniques listed with current instrumentation, making it widely adopted in the industry.
The main drawback of SPR is that it requires at least one component of the assay to be immobilised or captured onto a sensor surface, which is not always feasible. This could be due to interference from the chip or difficulties in keeping challenging protients active when captured.
Other technologies like MST, ITC, NMR, and FTSA get around this issue by being solutions but have their limitations. MST cannot generate kinetic data and requires separate assays to determine stoichiometry. Tag placement is also critical to the success of an assay, but there are situations where MST has proven to work when SPR cannot.
Ligand-observed NMR is highly reliable and can serve as a platform for extensive structural studies, albeit with limited applicability for high-affinity molecules. In contrast, ITC is the best way to gather energetic data for a project, delivering highly reliable results when executed correctly. Nevertheless, its throughput is notably lower than other techniques and requires care when setting up. It also has a very high reagent usage, particularly from a protein perspective, which often rules it out for anything other than the detailed characterisation of key examples.
These technologies can supply all the necessary data when applied appropriately, yet certain project constraints may limit the availability of these options. It is here that the scope of these technologies changes. For challenging targets, such as membrane proteins, or when working with covalent molecules, the most important thing to have is options, and it is here that a multi-technology approach to biophysics shows its strength.

Download your free eBook: Successful In Vitro Strategies for Drug Discovery
Learn more about optimal cascade design, hit-to-lead phase insights and lead optimization strategies.
The Power of Multiple Approaches
Despite having the best intentions and choosing a technology that should give the answers, there may be (and often is!) an unforeseen incompatibility between assay components and the desired equipment. While some issues can be addressed through assay optimization for the target, others, especially with more challenging targets, may be more fundamental. In such cases, biophysicists must carefully assess the tools at their disposal.
While everyone tends to have their preferred starting point, it’s essential to have a comprehensive understanding of the available and viable options. This approach can significantly enhance project effectiveness compared to sticking exclusively to one assay technology, even if it entails forgoing specific data types like kinetics. This dynamic approach is particularly suited to more difficult target classes, such as membrane proteins, helicases, and intrinsically disordered proteins (and more!). In these cases, casting a wide net from the outset often proves advantageous in kickstarting a project.
Does this approach always generate the correct data?
The vital parameters needed in early-stage projects, such as target engagement and affinity determination, can usually be obtained through multiple techniques, serving as crucial foundations for project initiation. As the potential therapeutics progress and increase in potency, and the understanding of target biology is improved, we find that other assay options become more viable to gain the required data.
There will always be exceptions to this, but at the very least, aligning the biophysical data generated on a project alongside other assays, such as activity or cellular assays, will help build confidence across the screening cascade, fill in gaps in understanding, and lead to better quality therapeutics as a result.
About the author
Dr Philip Addis is a highly experienced biophysicist and leads the biophysics group at Sygnature Discovery. Philip has over 10 years of biophysics experience in biotech and CROs, and has supported drug discovery projects for small, medium, and large molecules across a wide range of therapeutic targets.
