Why Free Energy Matters
Binding free energy quantifies how strongly a ligand interacts with a protein target. Accurate prediction of these energies is critical for ranking compounds and prioritizing synthesis in early drug discovery. One of the principal goals of any drug discovery project is to maximize affinity and selectivity against the desired target. Accurate prediction of protein-ligand binding free energies therefore plays a significant role in computational chemistry and computer-aided drug design.
With recent advances in force fields, sampling algorithms, and cloud-based GPU computing (GPU’s and cloud infrastructure), physics-based methods such as alchemical or relative binding free energy calculations have shown promise and have become powerful too ls for ranking and identifying the best compounds early in discovery.
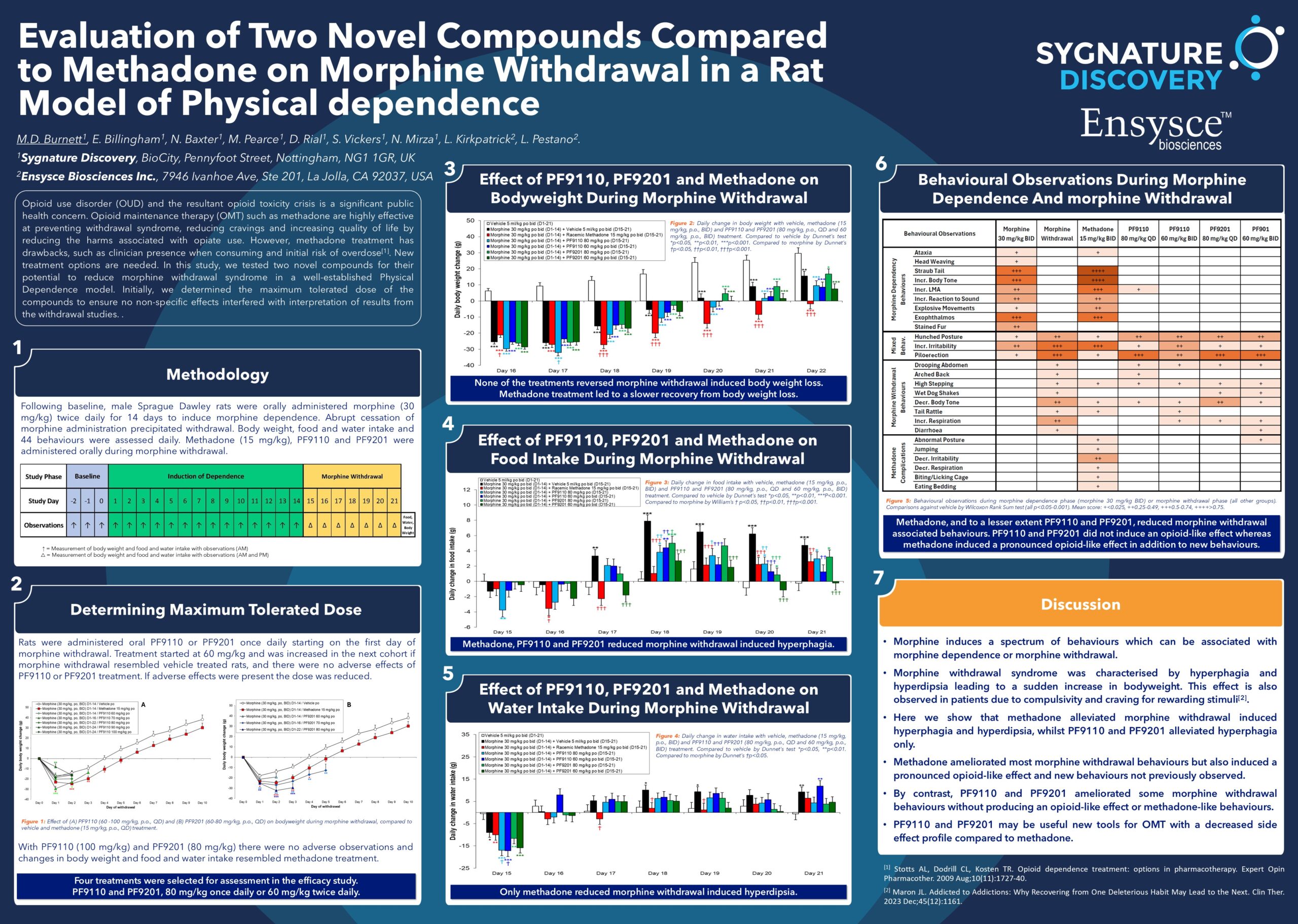
How NES Works
Relative alchemical binding free energy calculations involve simulations that interpolate interaction and internal energies between pairs of molecules. Data collected during these simulations estimate the difference in binding free energy between the two molecules. Among the most accurate and reliable predictions are Free Energy Perturbation (FEP) and NES.
We use OpenEye’s NES platform for predicting relative binding free energies of protein-ligand complexes with high accuracy and efficiency. Hosted on Orion’s scalable cloud infrastructure, NES leverages hundreds of CPUs and GPUs to deliver results faster than traditional techniques, significantly speeding up decision-making of compounds. OpenEye’s Orion also provides flexible edge-mapping algorithms such as OELOMAP, Starmap, Binary Starmap, and Multi Starmap tailored to your data and compound set. Together, these methodologies help you prioritize the most promising compounds to synthesize.
Why Choose Sygnature Discovery
Our team of computational and medicinal chemists bring extensive experience in understanding free energy calculations, including NES. We have proven successes in integrating these computational insights into real-world synthesis decisions.
We offer a fast, accurate way to predict ligand binding affinity so compounds for synthesis can be prioritized, saving significant costs and time.
Our Solutions
We’ve successfully applied NES to structurally enabled projects. In one lead optimization project against a kinase target, NES achieved strong correlation with experimental data (r2 = 0.76) and rank correlation of 0.71, validating its predictive power. Prospective studies confirmed NES as a reliable tool for filtering out low-potential compounds, saving time and cost.
Over the following year, NES was applied prospectively to more than 70 compounds across 13 NES runs representing 5 diverse chemical series. NES achieved a rank correlation of 0.62. When classifying compounds using a ΔG threshold of ≤ –10 kcal/mol, NES reac hed an accuracy of 0.75 and a Cohen’s kappa of 0.51. Except for one case, all high-affinity compounds, and many low-affinity compounds were correctly identified. These results demonstrate that NES can effectively filter out poor candidates with reasonable confidence, reducing both cost and experimental effort.
FEP vs. NES
Across multiple target studies, we compared the performance of FEP+ vs NES and found both the tools to perform with comparable accuracy.